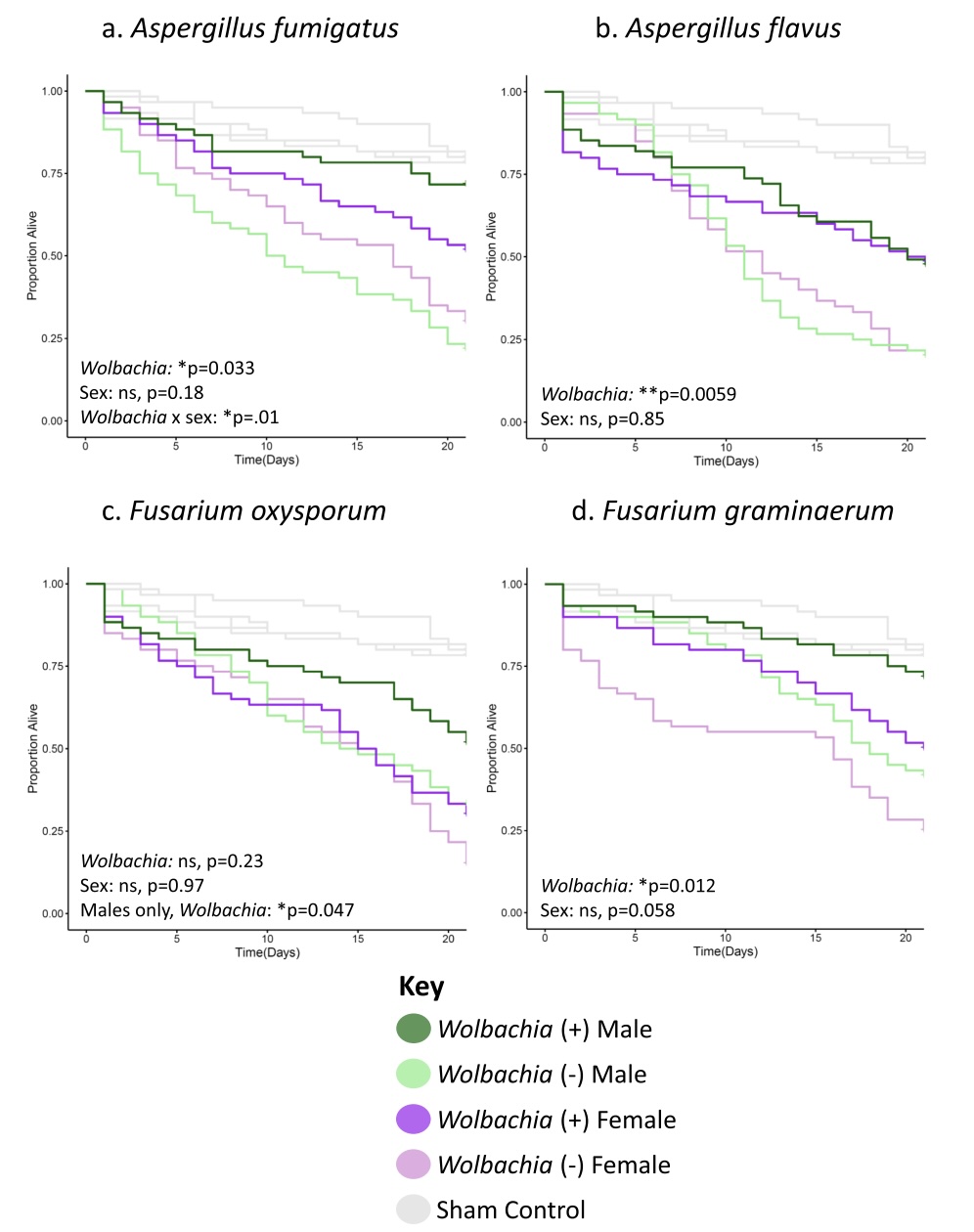
News
November 10th, 2025 - New lab photo - Fall 2025

Left to right(ish): Taiye, Nilanjan, Abby, Ashley, Sofia, Camila, Taylor, Rob, Kistie, Carly, Kervens, Vincent, Alexis, Todd, Lane, Anjali, Kaylie (missing: Demetrius, Xavion, Lily, Rohan)
November 7th, 2025 - Free State High School Women in STEM club visits the lab
We had the FSHS Women in STEM club visit the lab on October 22nd. The group, led by teacher Sara Abeita, saw various aspects of lab work. Kistie (technician) did a great job organizing the visit.
November 6th, 2025 - Avery Johnsmeyer did his whiteboard artistic interpretation of our lab's work. Impressive stuff.
November 1st, 2025 - Two new rotation students: Lane and Alexis
| Lane |
Alexis |
 |
 |
Lane Anaya and Alexis Michalski are new rotation students for the second rotation. Lane, who is returning to the lab after working with us as an undergraduate, will work on meiotic drive. Alexis, fresh off a Dalton lab rotation, will work on genetic variation in immune defense.
Lane Anaya (left) and Alexis Michalski (right) are new rotation students for the second rotation. Lane, who is returning to the lab after working with us as an undergraduate, will work on meiotic drive. Alexis, fresh off a Dalton lab rotation, will work on genetic variation in immune defense.
October 31st, 2025 - The Unckless Lab wins Haworth Haunt!

Every year Undergraduate Biology at KU holds the Haworth Haunt - a halloween door decorating competition. This year, the Unckless lab door won!
October 18, 2025 - Rotations, new people in the lab and a visitor from Denmark!

This fall we've had several new people join the lab (or at least visit us for a bit)...
- Todd Farmer is a postdoc working on a project that is a collaboration between the Kelly/Macdonald/Unckless labs
- Ashley Bentz is a technician working on that same project
- Tessa Eads is a 1st year graduate student in MB and was a rotation student during the first rotation. Lane Anaya and Alexis Michalski are also MB students and they will rotate during the second (and third) rotations
- We have several new undergraduates working in the lab: Lily O'Donnell, Xavion Smith and Kaylie Schroeder
- Sofie Ebsen is visiting from Aarhus University in Denmark to learn about the Drosophila innubila Nudivirus
August 21, 2025 - Welcome Camila Beraldo!

Camila Beraldo joined the lab as a postdoc. She recently completed her PhD at the University of Helsinki in Finland working with Anne Duplouy on symbiosis and the microbiome. She will work on evolutionary divergence in immune defense between species at KU.
July 19, 2025 - Jessie Perlmutter leaves to start her own lab at the University of Virginia
Jessie started at KU in March of 2020 after an incredibly successful graduate degree at Vanderbilt with Seth Bordenstein. She continued to do great things at KU including…
getting her own funding: an NSF PRFB and an NIH K99!
publishing: papers in PLOS Genetics, MBE, BMC Biology and more!
outreach and community building: Jessie founded the Kansas Postdoc Outreach Program (KPOP) and was a leader in KU Center for Genomics efforts including the annual Symposium.
mentoring: Jessie was an exceptional mentor to others in the lab serving as almost a second advisor and a good sounding board for me
the lab social organizer: she organized outings (Ren Fest), book clubs, the often disappointing lab trivia team, and more.
Jessie will be missed dearly, but we are looking forward to seeing what she does in her own lab. She is joining the Department of Biology at the University of Virginia and will work on various aspects of symbiosis, Wolbachia and immune defense. Good luck Jessie!
Research in the Unckless Lab

The overarching theme of the Unckless Lab research program is to understand the genomic and evolutionary consequences of genetic conflict at two different levels. Genetic conflict can be broadly defined as instances where different genomes or parts of genomes are at odds with each other and therefore attempt to manipulate each other for their own benefit. Between individuals, this conflict usually involves host/pathogen interaction. We focus on such host/pathogen interaction using the model species, Drosophila. This work involves the interactions between hosts and their bacterial pathogens with an emphasis on host antimicrobial peptide defenses. We also work on the evolution of host/virus interactions using Drosophila and a large, double-stranded DNA virus, the Drosophila innubila Nudivirus. The second level of genetic conflict is intragenomic conflict where different parts of the same genome (i.e. different chromosomes) compete for resources and transmission to the next generation. We study a particular case referred to as sex-ratio meiotic drive, where the X chromosome kills or disables sperm carrying the Y chromosome during sperm development.
For a long time, the mantra was that genes involved in immune defense were rapidly evolving by positive natural selection. This is because hosts and pathogens are involved in a coevolutionary arms race and as pathogens evolve stronger virulence or avoidance of the host’s immune defense, the host is then under evolutionary pressure to evolve new and/or stronger defenses. The process repeats ad nauseum resulting in rapid evolution in both host immune proteins and pathogens. Our work (and that of others) shows that these “Red Queen” coevolutionary dynamics are not universal. Some genes involved in immune defense evolve under a model of balancing selection. Furthermore, even when rapid evolution is identified in a particular pathway (antiviral RNAi), that pattern is not necessarily consistent among taxa. In addition to our work on the evolution of immune defense, we’ve worked to understand the evolutionary and genomic consequences of meiotic drive.
The evolution of immune peptides
Our work on the evolution of antimicrobial peptides (AMPs) began by accident when, as a postdoctoral researcher, I found that most of the genetic variation for immune defense against the Gram-negative pathogen, Providencia rettgeri, was associated with variation in Diptericin. Within a single population of Drosophila melanogaster, I found multiple null alleles and a nonsynonmous SNP all strongly associated with immune defense1. Shockingly, I found the same types of variation segregating (null alleles and the same two amino acids at the same position) in Drosophila simulans. Furthermore, the mutations had similar effects in both species: the serine allele conferred strong resistance, the arginine allele conferred weak resistance and flies carrying the null allele were mostly dead within 48 hours after infection. Both the null alleles and the nonsynonymous polymorphism were independently derived in the two species. We argued that the observed patterns were consistent with balancing selection and we used several approaches to show that, in general, antimicrobial peptides are evolving under a model consistent with balancing selection in several Drosophila species. Balancing selection was particularly surprising for AMPs because they are molecules that interact directly with pathogens. In summary, we found evidence that antimicrobial peptides do not evolve rapidly in an arms race with microbes, but instead evolve under balancing selection where multiple alleles are maintained adaptively. We’ve proposed two models for the maintenance of genetic variation in AMPs: a) autoimmunity where variants that are more efficient at inhibiting microbes are also more deleterious in the absence of infection and b) pathogen specificity where variants better able to fight pathogen A are less able to fight pathogen B. Both models would lead to balancing selection, but only the autoimmune model would explain the patterns of segregating null alleles.
Our work involves both in vivo (in the fly) and in vitro (MIC and functional assays). The in vivo work involves testing whether AMP variants protect against different pathogens during systemic infection and also exploring whether life history tradeoffs might explain the maintenance of variation in populations. We use CRISPR/Cas9 editing to alter AMP copy number, create null alleles, and importantly, create single nucleotide changes to alter amino acid residues. We’ve used the alleles created for Diptericin to confirm that the serine/arginine polymorphism causes a 40-50% difference in survival after infection with P. rettgeri. We’ve also found that the Diptericin allele influences the gut microbiota Our model is that one allele protects against systemic infection, but also reduces longevity in the absence of infection due to dysbiosis of the gut microbiota.
The in vitro work involves expressing or purifying AMP variants and exploring inhibition and mechanism of inhibition in standard microbiological and biochemical assays. As such, we’ve expressed and purified two variants of the AMP, Metchnikowin, and tested several biochemical properties as well as microbial inhibition against a series of pathogens. We’ve found slight differences in antimicrobial properties between the two variants for fungi and bacteria.
Host/virus coevolution
DNA viruses cause significant morbidity and mortality in humans and are fundamentally biologically different compared to better studied RNA viruses. They often have large (>100,000 bp) genomes, many (>50) open reading frames and often show high levels of recombination and complex replication cycles. To date, however, there are few model systems where both host and DNA virus are easily studied and manipulated in the laboratory. We discovered a DNA virus, the Drosophila innubila Nudivirus (DiNV) that naturally infects several Drosophila species, including the genetics workhorse, Drosophila melanogaster. The virus is common in natural populations and virulent – increasing mortality and decreasing female fecundity. Another group has characterized the related Kallithea virus in D. melanogaster.
One of the main goals in the lab is to establish DiNV as a model system for studying host/DNA virus coevolution so that we could then start to understand coevolution with hosts. We find these DNA viruses particularly fascinating because they are so different from other pathogens: high recombination rates compared to bacteria and most RNA viruses, low mutation rates compared to RNA viruses and many genes (100 or more). We took two approaches to developing the system.
First, we’ve worked to grow the virus as an experimentally tractable lab system. This involved growth in cell culture and in live flies. We are currently cloning the virus into a bacterial artificial chromosome (BAC) and will perform genetic manipulation of the virus in the BAC.
Second, we characterized molecular coevolution of host and virus. This began with a broad survey of the common large, double stranded DNA viruses (baculoviruses and nudiviruses) which indicated that a few genes (helicase, LEF4 and LEF5) show strong signs of adaptive evolution across the viral groups. We then sequenced the DiNV genome (155,000bp, 100 ORFs) and found signs of positive selection in similar genes (helicase and a duplication of the PIF protein, ODV-e56-2) as in the broader survey. PIF proteins are involved in infection in the gut and ODV-e56 is an ortholog of PIF5 which plays a role in host range.
As part of the Drosophila innubila genome project, we sequenced two outgroup species to look at patterns of molecular evolution. Our work resulted in what was the most contiguous published Drosophila genome (29.59Mbp N50). We performed a comparative analysis of evolutionary rates between D. innubila and D. melanogaster and found that there was a general correlation between rates of evolution, but rates of evolution in immune categories varied significantly among species. In D. melanogaster, genes involved the antiviral RNAi pathway are among the most rapidly evolving in the genome, but in D. innubila, they look very average. In contrast, in D. innubila, Toll pathway genes are very rapidly evolving, but they are average in D. melanogaster. This correlates with what we know about natural pathogens. D. melanogaster is infected by numerous RNA viruses that are controlled by the antiviral RNAi pathway. We’ve looked extensively and found almost no evidence of RNA viruses in D. innubila. However, D. innubila is plagued by DiNV (DNA virus) which seems to interact with the Toll pathway. These first several papers both established genomic tools and initial patterns of evolution that serve as a basis for ongoing and future work.
We then moved on to a large-scale population genomic survey of both D. innubila and DiNV, sequencing more than 400 individual wild-caught flies. In the virus, we found something completely unexpected. First, while hosts move freely between geographically isolated populations, the viruses are strongly structured. Second, there are two haplotypes segregating among the viruses and they differ 100-fold in viral titer. The high viral titer haplotype is the derived state and evolved independently in three different populations. What’s amazing about this high type is that it involves 11 almost perfectly linked SNPs spread throughout the 155kb circular viral genome – while other interspersed sites are freely recombining. We performed RNA-sequencing and found that those infected with the high type had higher expression of gp83, which inhibits host Toll signaling. Finally, we sequenced individuals from a distantly related host species that overlaps in geographic range as well as from a more closely related species with no overlap in host range. Both species segregate for both viral haplotypes, and in both species the “high” haplotype is associated with about 100-fold higher viral titer. Our work on host/virus coevolution begins to uncover the way in which specific pathogen pressure drives the molecular evolution of host species and how malleable viruses are in evolving optimal virulence in their hosts.
Moving forward, we would like to understand how these virus types are maintained in populations and why they don’t recombine. Have they experienced some sort of viral speciation event where even though they may recombine to form chimeras, those chimeras are significantly less fit or inviable and therefore don’t persist? Second, we’d like to understand host/virus evolutionary interactions in a host community sense. Since this virus infects several different species at different frequencies, how do fly community dynamics influence viral type abundance and how does viral type influence hosts communities?
Sex-ratio meiotic drive
Our other major area of empirical research is a genomic and population level analysis of the Drosophila affinis meiotic drive system. Sex-ratio males, those carrying a driving X chromosome, sire nearly all daughters because the driving X disables Y-bearing sperm. Important work by Robert Voelker and colleagues in the 1960s and 1970s found that a) XO males (those lacking a Y) are fertile, b) a driving X chromosome produces nearly all daughters when in XY males, but nearly all females when in XO males, c) the Y outcompetes the O in the absence of the driving X, and d) a second driving X is not suicidal when paired with the O, but drive is much weaker. We aim to elucidate the genomic basis of drive and monitor long-term population dynamics of the two drivers and the Y/O polymorphism.
Our main goals at this point are using genetic and molecular techniques to characterize the genes and mechanisms involved in meiotic drive and resistance to meiotic drive.
Theoretical work
We are interested in how conflict shapes ecological and evolutionary processes. Our theory work in this area has recently focused on sex-ratio meiotic drive and synthetic gene drive. To date we’ve looked at the effect of meiotic drive on population extinction, speciation and sex-chromosome evolution.